Another paper on the role of IAM
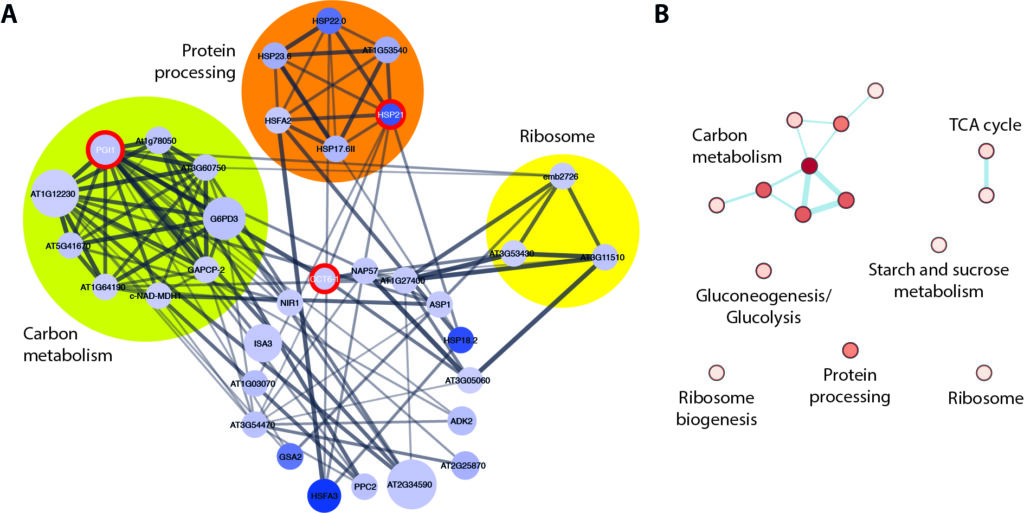
Network analysis of down-regulated differentially expressed genes (DEGs) in ami1 rty. (A) Extracted network for the three DEGs with the highest betweenness centrality. The central hubs CCT6-1, HSP21, and PGI1 are highlighted by red circles. The nodes are color coded according to the log2FC expression levels of the DEGs (white to blue), with dark blue marking the most repressed DEGs. The node size describes the false discovery rate (FDR) values, while the edge thickness reflects the betweenness score between the nodes. Densely connected nodes have been identified by using the Molecular Complex Detection (MCODE) clustering algorithm and are highlighted by colored circles. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the sub-network extracted for CCT6-1, HSP21, and PGI1. The color (white to red) gives account on the normalized enrichment score of the corresponding KEGG pathway.
Slowly but steadily we further our understanding on the possible role of IAM in plant growth and development. In our latest work on the ami1 rty double mutant, which was recently published in the International Journal of Molecular Sciences, we analysed the embryo phenotype of the mutant and comprehensively investigated the transcriptional differences in this line in comparison to wild-type Arabidopsis. It turned out that the accumulation of IAM, along with a profound impact on ribosome-biogenesis, triggers the repression of a subset of transcription factors controlling plant growth. So happy to see another great part of the PhD thesis of Beatriz finally published.
Sánchez-Parra, B. et al. 2021 Int. J. Mol. Sci.
The major auxin, indole-3-acetic acid (IAA), is associated with a plethora of growth and developmental processes including embryo development, expansion growth, cambial activity, and the induction of lateral root growth. Accumulation of the auxin precursor indole-3-acetamide (IAM) induces stress related processes by stimulating abscisic acid (ABA) biosynthesis. How IAM signalling is controlled is, at present, unclear. Here, we characterise the ami1 rooty double mutant, that we initially generated to study the metabolic and phenotypic consequences of a simultaneous genetic blockade of the indole glucosinolate and IAM pathways in Arabidopsis thaliana. Our mass spectrometric analyses of the mutant revealed that the combination of the two mutations is not sufficient to fully prevent the conversion of IAM to IAA. The detected strong accumulation of IAM was, however, recognised to substantially impair seed development. We further show by genome-wide expression studies that the double mutant is broadly affected in its translational capacity, and that a small number of plant growth regulating transcriptional circuits are repressed by the high IAM content in the seed. In accordance with the previously described growth reduction in response to elevated IAM levels, our data support the hypothesis that IAM is a growth repressing counterpart to IAA.


